Metabolomics
NMR is one of the most important techniques for metabolomics studies. At CERMAX we have a long track record of metabolomics studies using a large variety of biological materials like whole cell extracts, selected pieces of tissues (using the HR-MAS technique), or different body fluids like blood, plasma, sweat, urine or tears. Here are two recent examples:
Discovery of serum biomarkers for diagnosis of tuberculosis by NMR metabolomics including cross‑validation with a second cohort
Tuberculosis (TB) is a disease with worldwide presence and a major cause of death in several developing countries. Current diagnostic methodologies often lack specificity and sensitivity, whereas a long time is needed to obtain a conclusive result. In an effort to develop better diagnostic methods, this study aimed at the discovery of a biomarker signature for Tb diagnosis using a NMR‑based metabolomics approach. In this study, we acquired 1H NMR spectra of blood serum samples of groups of healthy subjects, individuals with latent TB and of patients with pulmonary and extra‑pulmonary TB. The resulting data were treated with uni- and multivariate statistical analysis. Six metabolites (inosine, hypoxanthine, mannose, asparagine, aspartate and glutamate) were validated by an independent cohort, all of them related with metabolic processes described as associated with TB infection. The findings of the study are according with the WHO Target Product Profile recommendations for a triage test to rule‑out active TB.
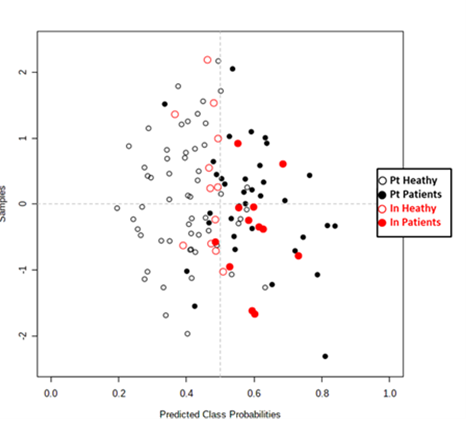
Six serum metabolites discovered in the Portuguese cohort predict correctly 92% of the samples from the Indian cohort . Prediction of the samples from the Indian cohort using the ROC curve created with the Portuguese cohort considering hypoxanthine, asparagine, mannose, aspartate, glutamate and inosine levels.
Reference:
A. Albors-Vaquer, A. Rizvi, Manolis Matzapetakis, Pedro Lamosa, Ana V. Coelho, A. B. Patel, S. C. Mande, S. Gaddam, António Pineda-Lucena, S. Banerjee e Leonor Puchades-Carrasco. (2020) Active and prospective latent tuberculosis are associated with different metabolomic profiles: Clinical potential for the identification of rapid and non-invasive biomarkers. Emerging Microbes & Infections. 9:1131-9. https://doi.org/10.1080/22221751.2020.1760734
Absence of HIF-1α increases susceptibility to Leishmania donovani infection and increased acetate uptake.
Hypoxia-inducible factor-1 alpha (HIF-1α) is considered a global regulator of cellular metabolism and innate immune cell functions. Intracellular pathogens such as Leishmania have been reported to manipulate host cell metabolism. Myeloid cells from myeloid-restricted HIF-1α-deficient mice and individuals with loss-of-function HIF1A gene polymorphisms are more susceptible to L. donovani infection through increased lipogenesis. Using 2-13C-acetate, it was possible to observe by 13C-NMR and 1H,13C-HSQC that increase uptake of acetate and its incorporation on the lipid fraction of infected HIF-1α−/− macrophages, suggesting acetate uptake as a result of enhanced fatty acid synthesis in infected HIF-1α−/− macrophages.
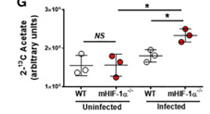
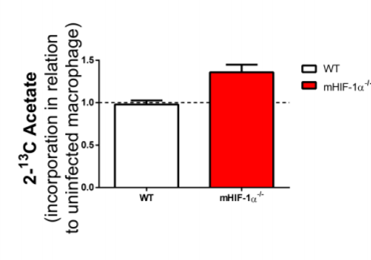
Increased incorporation of 13C-acetate on the lipid fraction of L. donovani infected HIF-1a-/-. Intracellular accumulation of the 13C label that was incorporated in 2-13C-labeled acetate was determined in lysates from uninfected and L. donovani-infected WT or HIF1α−/− peritoneal macrophages upon 24 h by NMR (right panel). WT and HIF-1α -/- macrophages were infected with L. donovani for three days following subsequent 24 hours incubation in the presence of 13C-labeled acetate (left panel).
Reference:
Mesquita I, Ferreira C, Moreira D, Kluck GEG, Barbosa AM, Torrado E, Dinis-Oliveira RJ, Gonçalves LG, Beauparlant CJ, Droit A, Berod L, Sparwasser T, Bodhale N, Saha B, Rodrigues F, Cunha C, Carvalho A, Castro AG, Estaquier J, Silvestre R (2020) The Absence of HIF-1α Increases Susceptibility to Leishmania donovani Infection via Activation of BNIP3/mTOR/SREBP-1c Axis. Cell Rep, 30:4052-64.e7. doi: 10.1016/j.celrep.2020.02.098.
