Structural Biology
The structure and function of FhuF, a ferric-siderophore reductase from E.coli
FhuF is the only bacterial ferric-siderophore reductase ever isolated given the instability found on these type of proteins. It contains an atypical 2Fe-2S with the motif Cys-Cys-X10-Cys-X2-Cys and although the function of FhuF as a ferric-siderophore reductase was already established, there is presently no knowledge regarding its structure and how this atypical 2Fe-2S cluster operates to mediate ferric-siderophore reduction.
Given the difficulty in crystallizing FhuF, NMR spectroscopy was explored as a way of studying the structure of this protein, mapping the interactions between FhuF and redox partners and also for studying the structure and electronic properties of the atypical 2Fe-2S cluster.
The TROSY-HSQC spectrum of the U-15N/13C labelled protein misses only 15 backbone peaks out of 235 expected, indicating that it is possible to proceed with the resonance assignment. Also, paramagnetic studies in both oxidized and reduced states of FhuF were performed revealing the unique structural and electronic properties of the 2Fe2S active site.
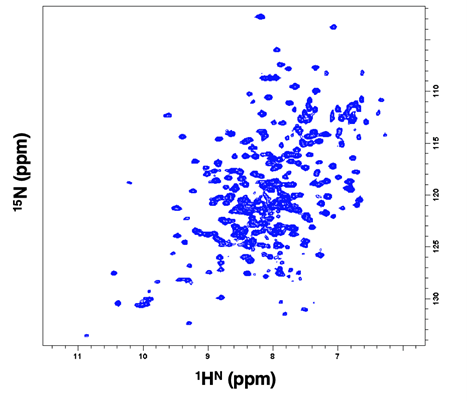
Fig.1- 2D 1H15N TROSY-HSQC spectrum of U- 15N/13C-labeled FhuF collected at 305 K on a Bruker Avance III 800 MHz spectrometer with cryoprobe, showing a good dispersion of peaks.
Reference:
Trindade IB, Hernandez G, Lebègue E, Barrière F, Cordeiro T, Piccioli M, Louro RO, Conjuring up a ghost: Structural and functional characterization of FhuF, a ferric siderophore reductase from E. coli, J Biol Inorg Chem (2021) DOI: 10.1007/s00775-021-01854-y
Exopolysaccharide structure by NMR
Exopolysaccharides (EPSs) are of paramount importance in texture and taste of fermented foods, particularly in the dairy industry in the use of Lactic Acid Bacteria (LAB) for the production of cheeses and yogurts. The organoleptic properties of EPSs are derived from its structures (monomer composition, degree of branching and glycoside linkage patterns). Even closely related strains may have very different EPS structures. Experimental structure elucidation of EPS is time-consuming and expensive. So, the possibility to derive EPS structure from genomic information would be highly appealing. Here, we have investigated the structures of EPSs from 12 strains of S. thermophilus whose genome information is available, that revealed 6 novel repeat unit structures not previously observed, and derived a procedure to putatively assigned glycosyltransferases to one or more monomers in the repeat units. This is a first step towards predicting EPS structures from genomic information.
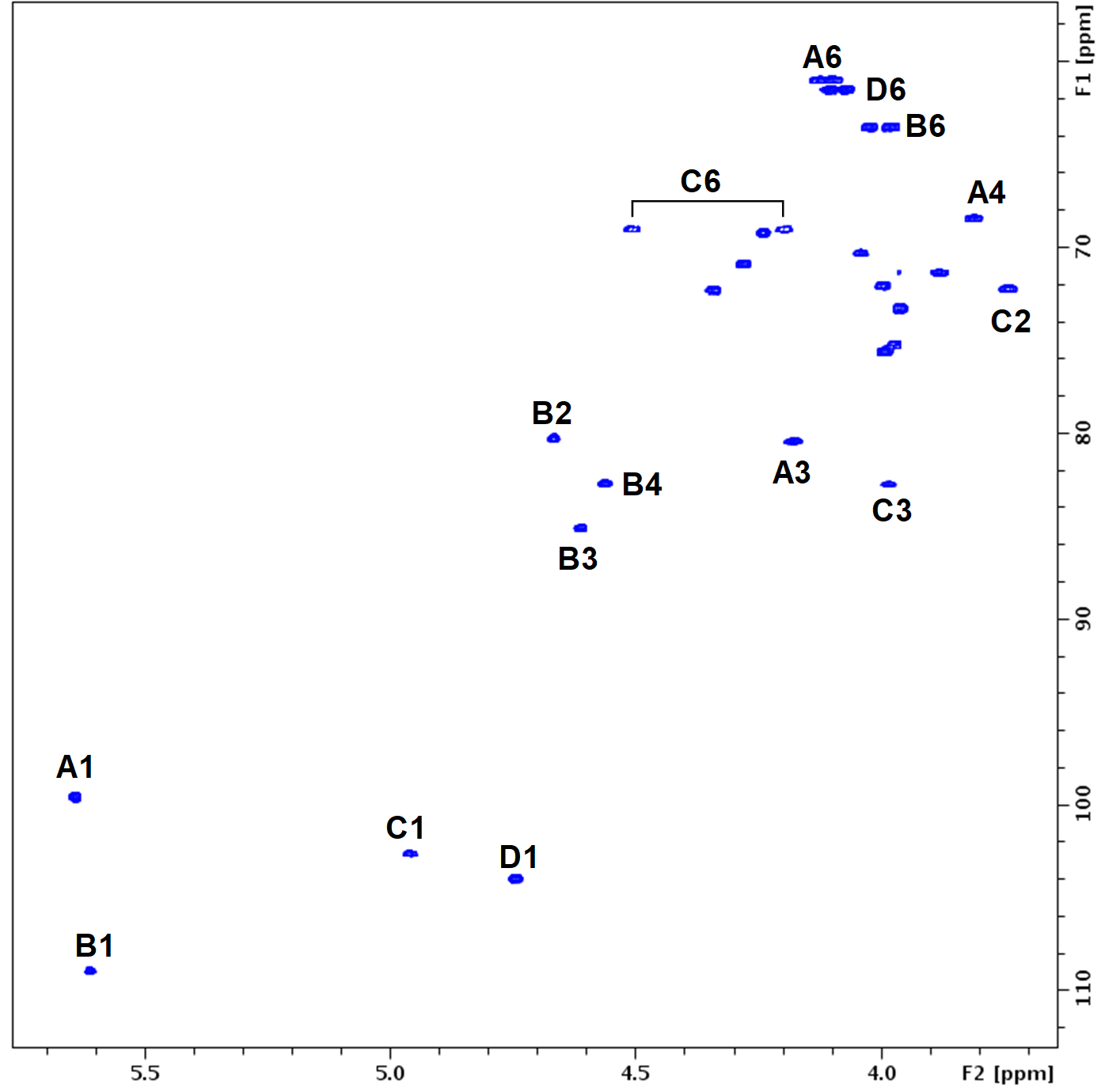
1H-13C HSQC acquired at the AVANCE III 800 spectrometer of the exoplysaccharide from S. thermoplilus strain CH9204
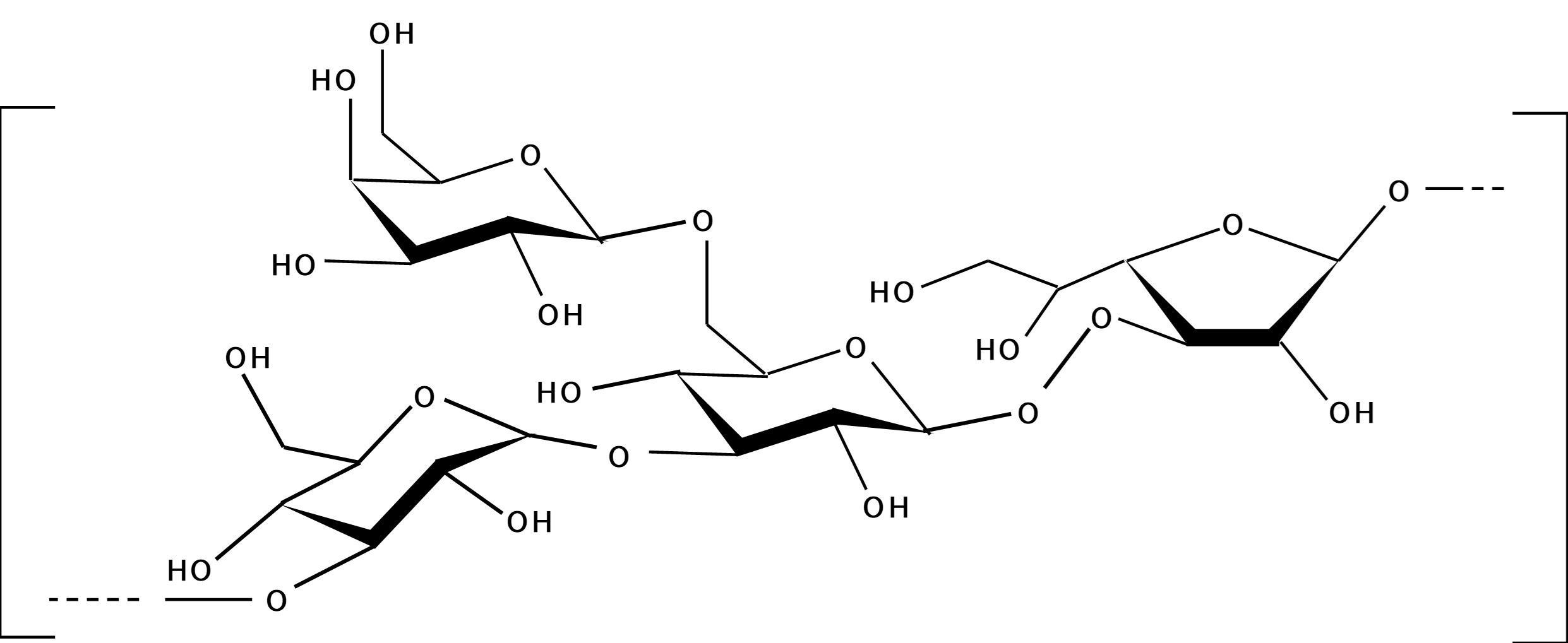
Example of a structure of the repeating unit of the exoplysaccharide from S. thermoplilus strain CH9204
